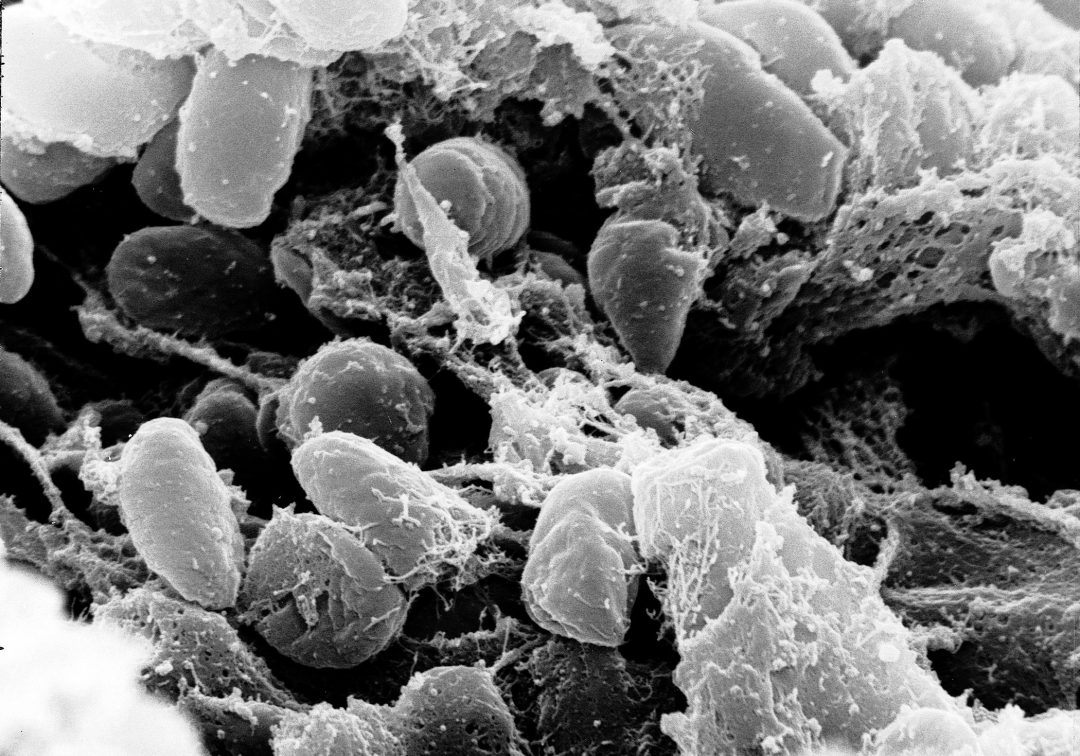
A team of researchers which includes members of the NEOMATRIX network developed a fully automated ancient metagenomics pipeline. This promising method will address some important challenges in the area of ancient metagenomics, such as a relatively high false-positive discovery rate, where exotic microbes, animals, plants, fish etc. can often be found where they are not supposed to be present. aMeta’s databases are ready for download, and the pipeline is open-source on GitHub.
The article is authored by Zoé Pochon, Nora Bergfeldt, Emrah Kırdök, Mário Vicente, Thijessen Naidoo, Tom van der Valk, N. Ezgi Altınışık, Maja Krzewińska, Love Dalén, Anders Götherström, Claudio Mirabello, Per Unneberg, and Nikolay Oskolkov.